
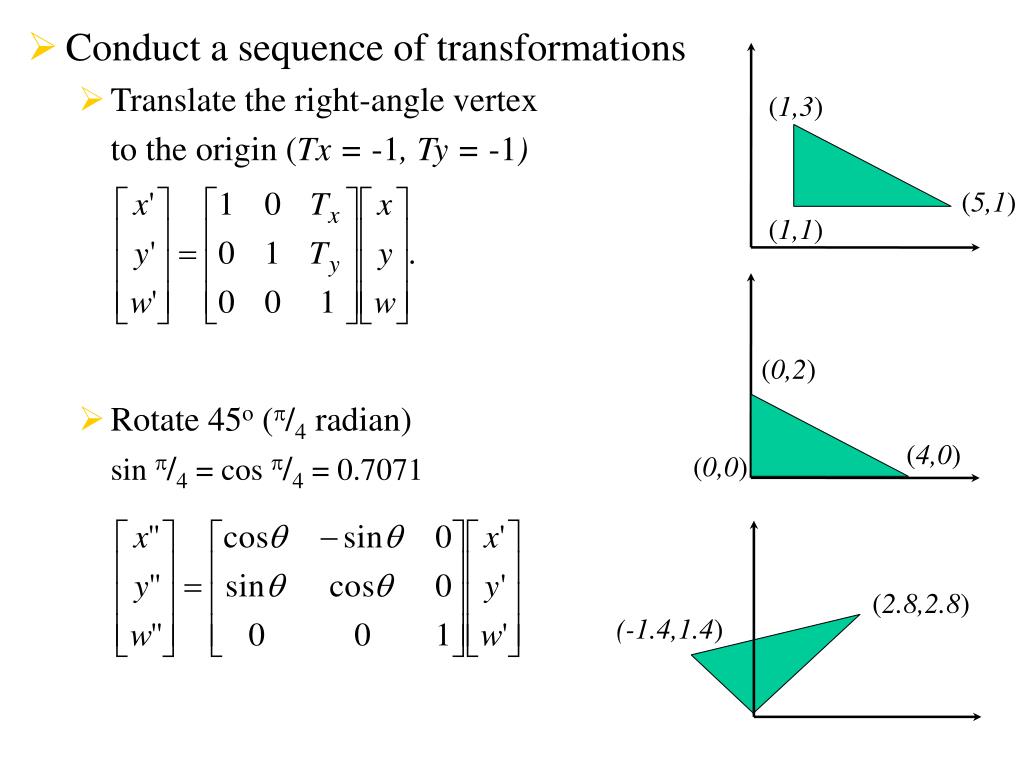
Equally, its content may be rewritten in other formats like the most elementary structure format. The later format is one many other programs accept as input. A script like codcif2sdf, part of the cod-tools collection, however is able to recognize the described symmetries of the unit cell acting on the molecule, and to join the molecular fragments into a. This is because the threefold symmetry axis of the molecule aligns with a threefold axis of the unit cell. cif describes only coordinate tuples for seven atom as such, which is exactly a third of the molecule. The initial display (of the asymmetric unit) by the COD and the complete molecular structure are provided below: This answer aims to complement the previous answers from the perspective not wanting to write new programs.Įntry COD 2102215 of the COD about 2,4,6-trimethoxy-1,3,5-triazine serves as an example. On the other hand, on first glance by visual inspection, the model might appear incomplete. cif file need not to contain as many explicit coordinate tuples as atoms in the molecule. However on occasion, the symmetry of the unit cell is higher and the molecule of interest is on a special position, for example a centre of inversion. Without doubt, the previously provided answers will work well if the crystallographic model is a description in space group triclinic $P1$. You should be able to confirm that the arrays Y and Y_ are the same.
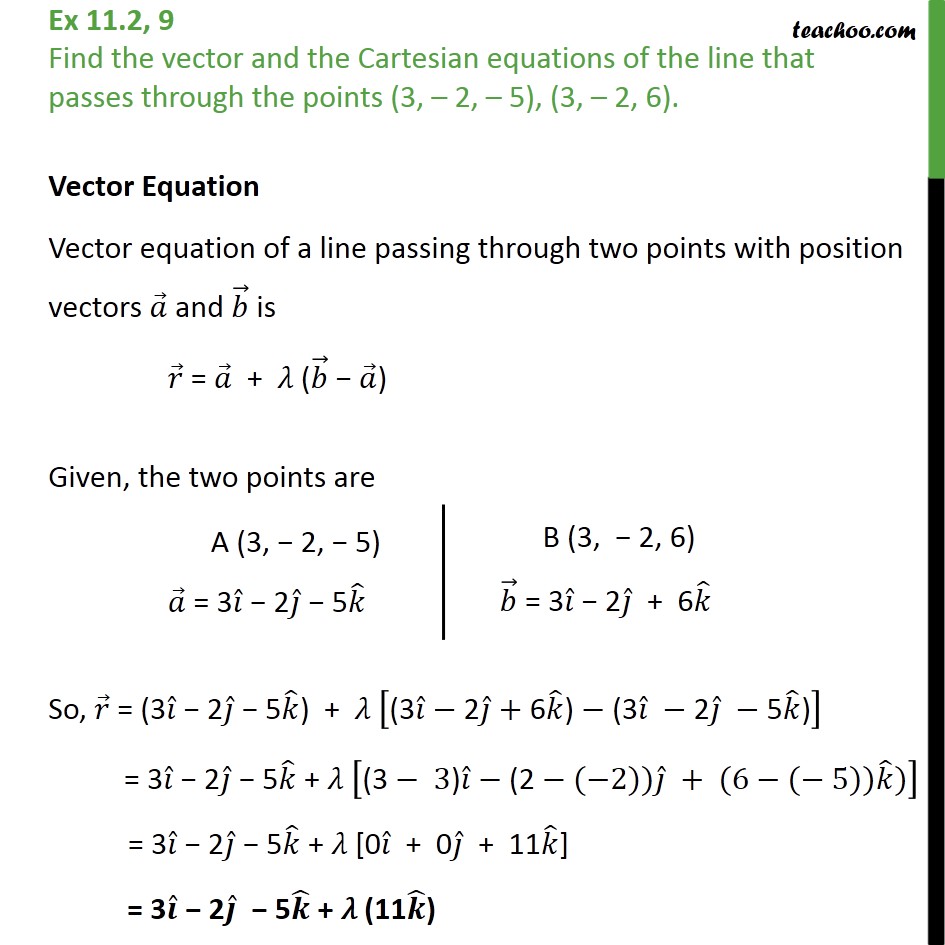
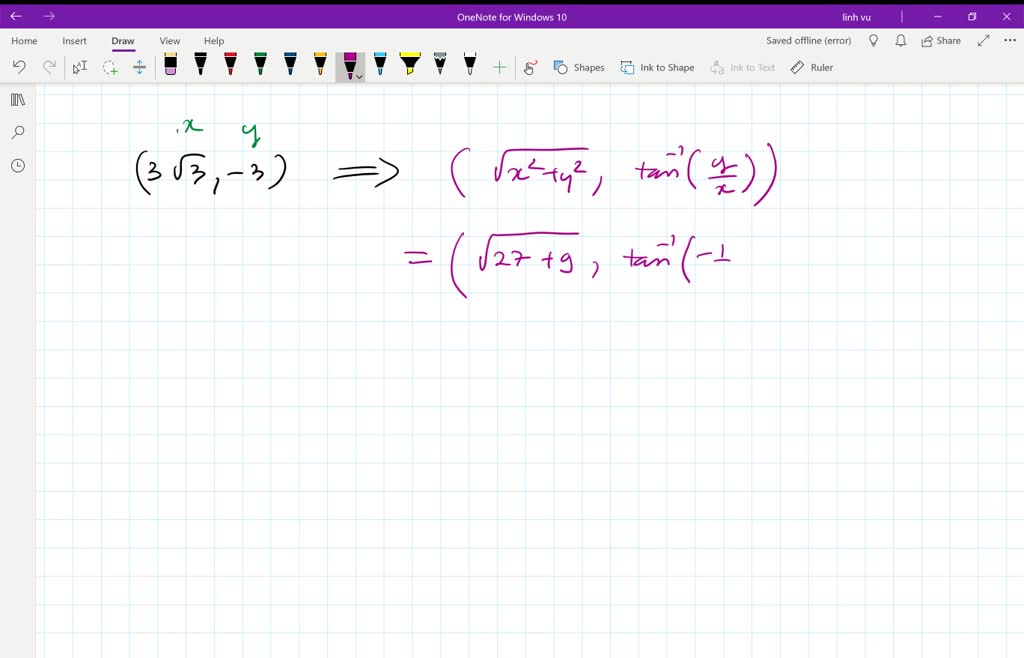
# Perform the inverse operation to get fractional coordinates # A random set of fractional coordinates stored row-wise # Build the matrix of lattice vectors stored column-wise In Python you can do the following: import numpy as np To get the corresponding set of atomic positions in cartesian coordinates denoted as $X$ (also an $m\times 3$ matrix), you can use the following equation:Īnd similarly to convert a set of cartesian coordinates to fractional coordinates you can use the inverse relation: So, now if you have a set of $m$ atomic positions in fractional coordinates, you can organize them into an $m\times 3$ matrix denoted as $Y$. To convert from cartesian coordinates to fractional coordinates, you can just trivially invert this equation to get: $$ M=\begin$, we can convert between the two using the following equation: This is derived in most textbooks on crystallography, such as McKie & McKie 'Essentials of Crystallography'. %chk=D:\!HNGH\aug-cc-pwCVTZ\CCSDT XeH2 freq.You have to use a matrix to convert. But how do I fix this? I can't really use the results obtained untill the error is dealt with, can I? Conversion from Z-matrix to cartesian coordinates failed:'Īpparently, Gaussian doesn't deal well with linear fragments and this molecule is linear. It reads 'Error on Z-matrix line number 3: angle Alpha is outside the valid range of 0 to 180. But the output file doesn't contain the fragment that should say that all 4 convergence criteria are satisfied and that a stationary point is located.
What's even weirder is that it seems to happen right after everything is already calculated (all the energies and corrections). This error occurs in the frequency calculation.


 0 kommentar(er)
0 kommentar(er)
